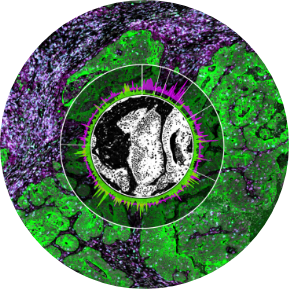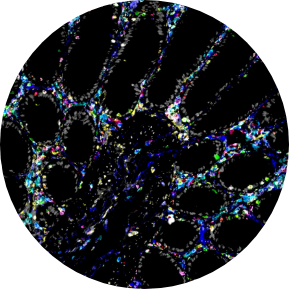
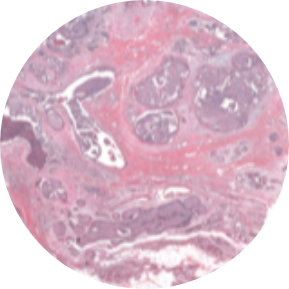
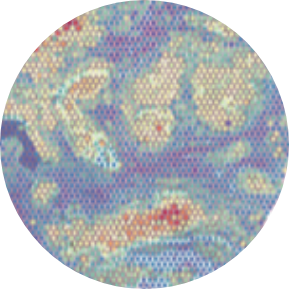

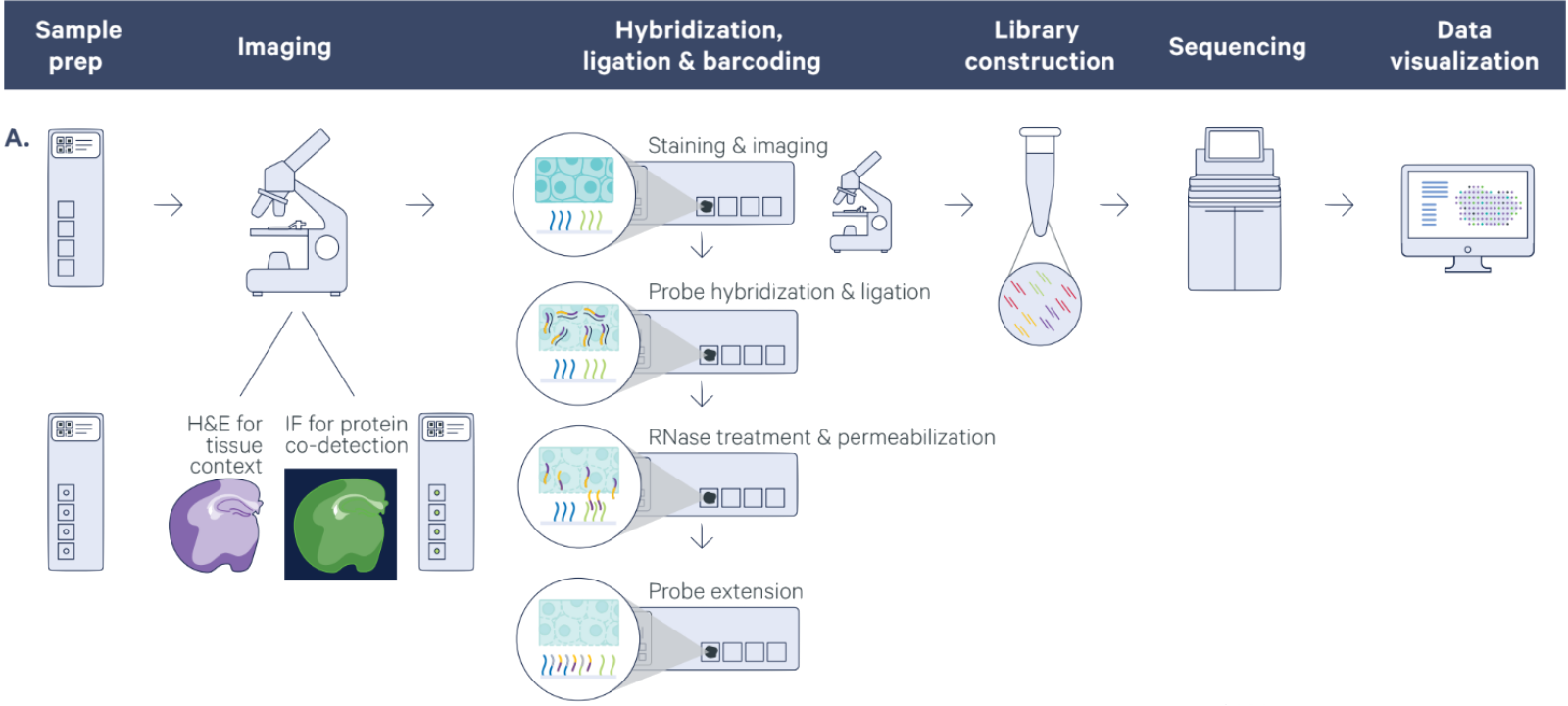







The use of 10x Genomics technology permits researchers to conduct high-resolution genomic analysis, which facilitates the examination of intricate biological systems and the discovery of previously concealed information regarding cellular behavior and function.
With the provision of advanced tools and technologies, 10x Genomics seeks to expedite scientific breakthroughs and reveal fresh perspectives on human health, disease mechanisms, and other biological phenomena.